DNA programming is the DNA-counterpart of computer programming. As illustrated in figure 3, the basic computer programming cycle is to modify an existing program, test the modified program, and iterate until the desired behavior is obtained.
Similarly, the DNA programming cycle is to modify a DNA molecule, test its resulting behavior, and iterate until the goal (which is either understanding the behavior or improving it) is achieved. One key difference between the two is that unlike computer programming, our understanding of DNA as programming language is very far from being perfect, and
therefore trial and error are the norm rather than the exception in DNA-based research and development. Hence DNA programming is more efficient if multiple variants of a DNA program, also called a DNA library, are created and tested in parallel, rather then creating and testing just one program at a time. Hence the basic DNA programming cycle, when operating in full steam, takes the best DNA programs from the previous cycle, uses them as a basis for creating a new set of DNA programs, tests them, and iterates until the goal is achieved.
The biotechnology revolution has, to a large extent, been held back by its notoriously prolonged R&D cycle compared to the computer programming cycle. A CAD/CAM technology for DNA which will bring word processor ease to DNA processing and thus support rapid DNA programming will revolutionize biotechnology by shortening the R&D cycle of DNA-based
applications. This can only be accomplished by concerting the development of complex, multi layered technologies which integrate expertise from fields as varied as algorithmics, software engineering, biotechnology, robotics and chemistry. These are only now starting to emerge as feasible.
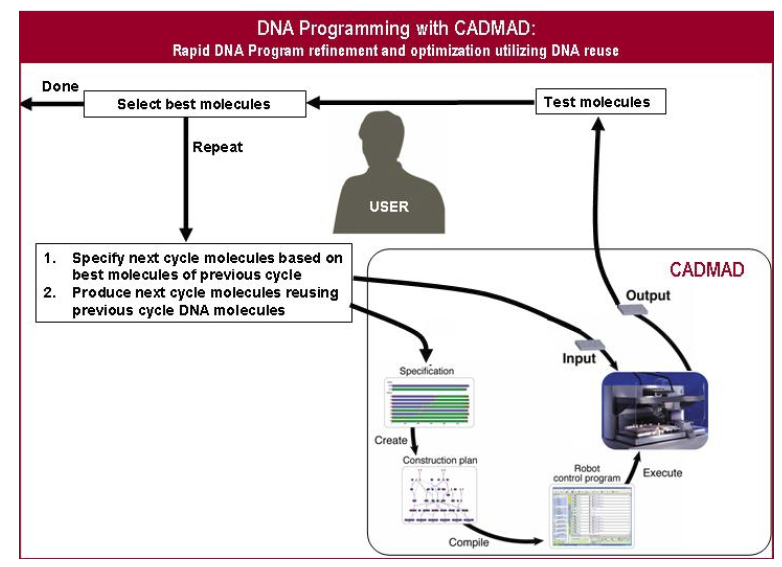
Furthermore, Polymerase Chain Reaction (PCR) is the DNA-equivalent of Gutenberg's movable type printing, both allowing large-scale replication of a piece of text. De novo DNA synthesis is the DNA-equivalent of mechanical typesetting; both ease the setting of text for replication. What is the DNA-equivalent of the word processor? Word processing was rapidly adopted as a replacement for the typewriter when users had discovered its revolutionary advantages in document creation, editing, formatting and saving. While the electronic representation of text in computers allows the processing of text within a simple unified framework, DNA processing — the creation of variations and combinations of existing DNA - is performed by biology labs daily using a plethora of unrelated manual labor-intensive methods. As a result, so far no universal method for DNA processing has been proposed and, consequently, no engineering discipline that further utilizes the processed DNA has emerged. The CADMAD technological platform aims to deliver a revolution in DNA processing analogous to the revolution text editing underwent with the introduction of electronic text editors.
| The Challenge | Text | DNA |
|---|---|---|
| Automated Large-scale Replication |  Movable type printing, Gutenberg, 1439 Movable type printing, Gutenberg, 1439 |
 PCR: Polymerase Chain Reaction, Mullis, 1983 PCR: Polymerase Chain Reaction, Mullis, 1983 |
| Setting a text/DNA for replication |  Mechanical typesetting Machines (Linotype, Monotype), 1880's Mechanical typesetting Machines (Linotype, Monotype), 1880's |
DNA synthesis factories, 2000's
|
| Text and DNA proccessing |  Word processors on personal computers, 1980's Word processors on personal computers, 1980's |
CADMAD
|
CADMAD aims to be the DNA-equivalent of the word processor as an essential tool in rapid DNA program development. 




